3D imaging for serial section by using automation image acquisition software.
- Abstract number
- 692
- Event
- European Microscopy Congress 2020
- DOI
- 10.22443/rms.emc2020.692
- Corresponding Email
- [email protected]
- Session
- LST.4 - Volume Scanning Electron Microscopy in life sciences
- Authors
- Hiroyuki Chiba (1), Takashi Shidara (1), Shigeaki Tachibana (1), Shigeru Kawamata (1), Mami Konomi (1)
- Affiliations
-
1. Hitachi High-Tech Corporation
- Keywords
ArrayTomography
- Abstract text
Due to recent improving the observation performance of low-acceleration voltage backscattered electron images by high-resolution FE-SEM, it has become possible to obtain images similar to TEM images at observing biological samples embedded in a resin.
Especially in the section observation using SEM, unlike TEM observation in which the section is mounted on a grid, the section can be mounted on a substrate such as a microscope slide and observed.
This not only reduces the difficulty of sample preparation, but also facilitates wide-area observation, is applied to biological tissue and whole cell observation increasingly [1].
Furthermore, in addition to understanding large-area structures, SEM has been applied to study for the three-dimensional structure by observing new resin surface obtained by repeatedly cutting resin embedded samples or by observing its serial sections.
Array Tomography [2] is one of the methods for the three-dimensional reconstruction by observing and stacking the same region of each section in the serial sections.
Compared with the FIB-SEM method and the serial block face SEM method, which are the three-dimensional reconstruction methods, Array Tomography has advantages such as being able to re-observing, and being able to observe a wide area of 500 µm2 or more.
However, in image acquisitions, since a large number of ultrathin sections are arranged on a substrate for observation, it is necessary to perform searching a field of view to specify the position of each sections. Furthermore, it is necessary to find the same target position in each sections, and to match the imaging field of view. Such enormous tasks of repeating these operations with several hundred sections has been a major problem.
In this study, we report the development of the assist software which simplify the tasks of finding the same target position in each sections and matching the field of view for the Array Tomography method, which has been an issue so far.
The developed software can execute various automatic adjustment functions for acquiring images with highly stabilized. Once area of interest defined and run, images from each sections are automatically acquired with adjusted SEM parameter such as focus and brightness/contrast.
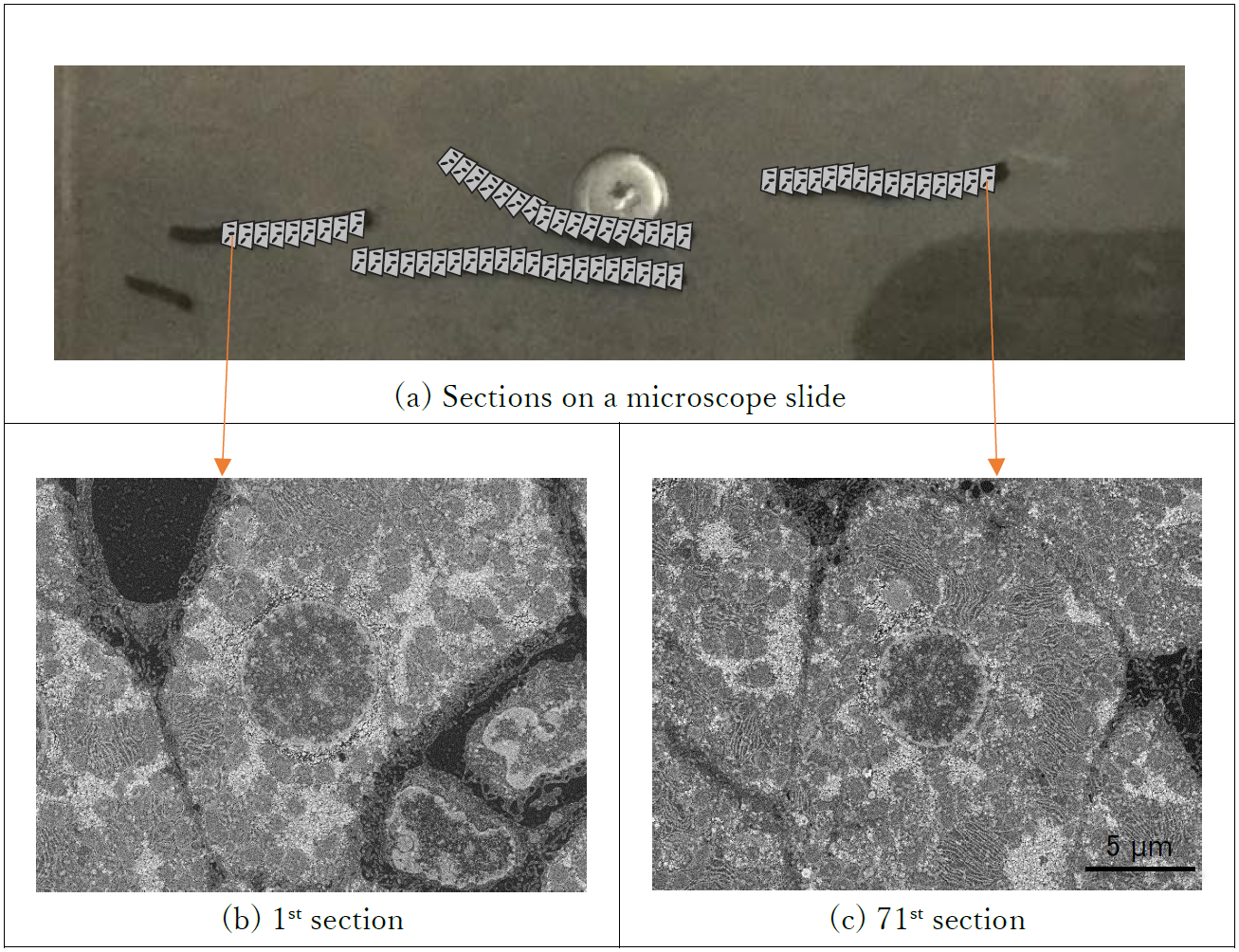
FIG. 1 shows the results of actual automatic imaging. It was confirmed that the focus, brightness, and contrast were adjusted by the automatic adjustment function in each sections from first to 71st images of the section 23.7 mm apart.
In addition, this software is implemented image recognition technique which compensates the position aberration of each section caused from deformation and/or curved line of serial section. This enables accurate imaging at the same position in each sections precisely.
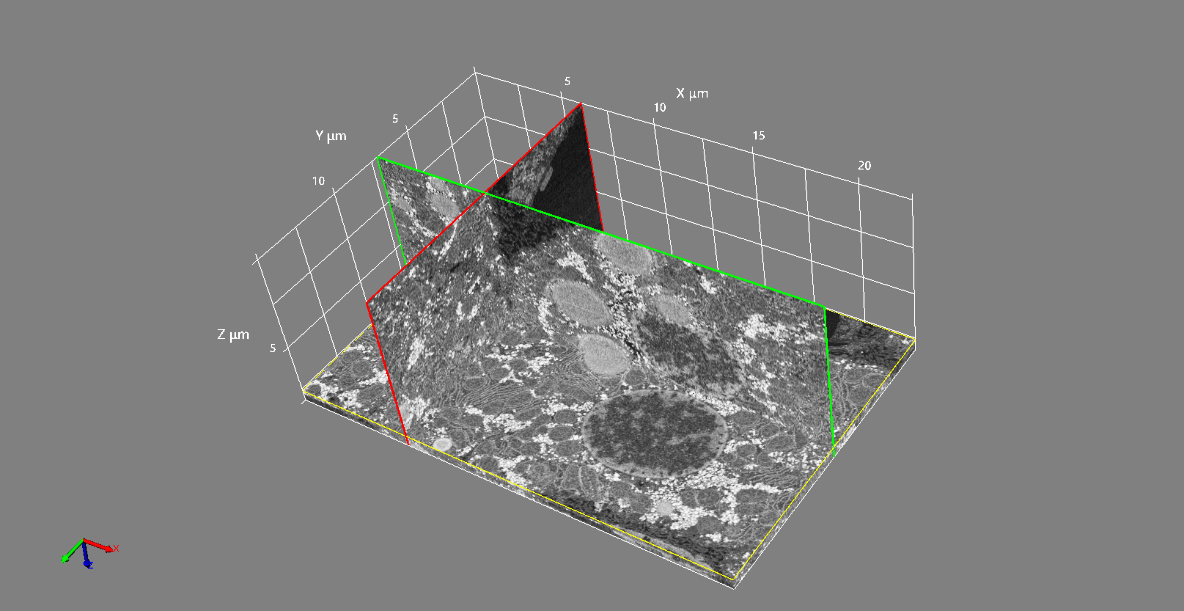
FIG. 2 shows a three-dimensional reconstructed data using 100 sections of image data automatically acquired using this software. We succeeded in acquiring a three-dimensional reconstructed data that can make understand the three-dimensional structure of rat hepatocyte cells.
As described above, the development of this system can solve the problem that searching a field of view is required and the work is enormous compared to the FIB SEM method and the serial block face SEM method. And this will contribute to other developed application in Array Tomography method furthermore.
Fig 1. Comparison of automatic acquired images between first and 71th section image in serial sections.
Fig 2. 3D reconstruction from serial sections of a rat hepatocyte
Acknowledgements sample was provided by Dr. Daisuke Koga of Asahikawa Medical. University, JAPAN
- References
1) Murvey T., Scanning Electron Microsc., 1: 43 (1974)
2) Micheva K.D. and Smith S.J., Neuron., 55: 25 (2007)