Bringing open microscopy to the Web of Things
- Abstract number
- 966
- Event
- European Microscopy Congress 2020
- DOI
- 10.22443/rms.emc2020.966
- Corresponding Email
- [email protected]
- Session
- DHA.3 - Machine assisted acquisition and analysis of microscopy data
- Authors
- Dr Joel Collins (1), Joram Mduda (2), Dr Julian Stirling (1), Dr Richard Bowman (1)
- Affiliations
-
1. Centre for Photonics and Photonic Materials, Department of Physics, University of Bath
2. Ifakara Health Institute
- Keywords
open source, 3d printed, networked, IoT
- Abstract text
The OpenFlexure Microscope is an open-source, 3D-printed motorised microscope (Figure 1) targeted towards medical applications in sub-Saharan Africa [1]. We have worked with local innovation hubs, research institutions, and clinics to develop control software that encourages contributions across all communities, from school pupils to professors.
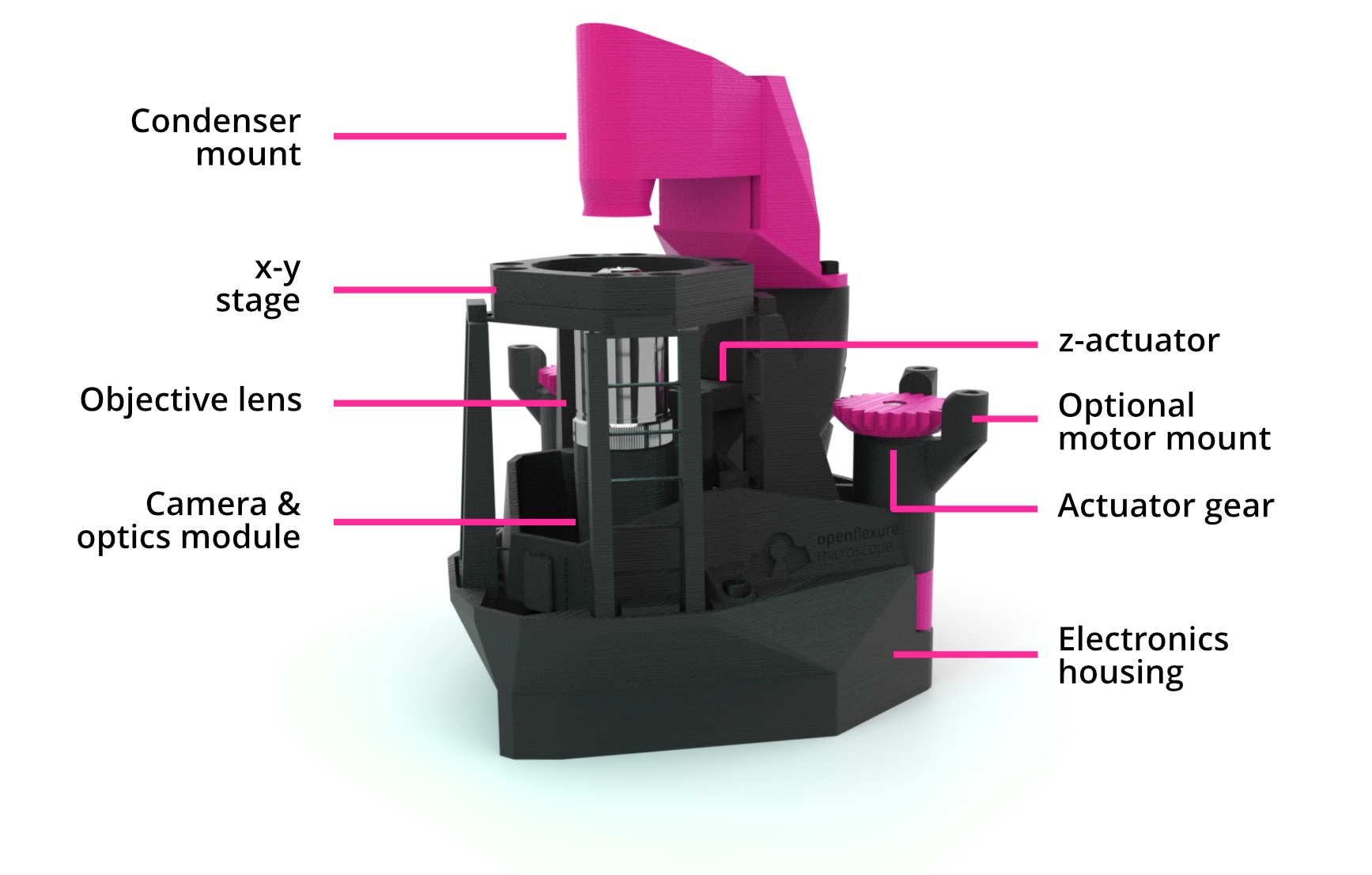
Figure 1 Overview of the OpenFlexure Microscope design (adapted from reference [1]). The condenser mount houses an illumination LED and a condenser lens. The optics module sits below the stage and houses an objective lens, tube lens, and camera, attaching to an actuator providing variable focus. Both the optics module and sample stage are controlled by gears at the back of the microscope, driven by stepper motors. A detachable electronics housing stores optional electronic parts, such as a motor controller and a Raspberry Pi microcomputer for automation.
The OpenFlexure Microscope has applications ranging from a low-cost educational tool all the way up to a research-grade laboratory instrument. Previously, everyone used their own set of hastily written control scripts, leading to duplicated effort and a user interface that was difficult for non-programmers to use. In response, we have rebuilt our control software from the ground up, with our collaborators in Cambridge and Tanzania. By introducing a new common core, a comprehensive plugin system, and a standard web API, we have been able to draw on the expertise of a diverse community to create a uniquely customisable software stack for controlling the microscope in any setting.
Our work has particularly focused on low-cost Malaria diagnosis, allowing automated analysis on a network of microscopes. Where trained microscopists are in short supply, this scalability helps existing technicians diagnose more patients, and facilitates better training. Our developments have also enabled school pupils to develop microscope control scripts as part of outreach projects and will soon see the microscope used in university-level teaching labs.
Open-source hardware is poised to revolutionise countless industries such as research, local manufacturing, and education. However, developing scientific instrumentation poses a unique set of challenges revolving around synchronisation, data transmission, and resource management. Many of these challenges have solutions found outside of scientific research, in Web of Things standards and technologies. Our work has adapted a range of these standards to create a modern, connected cluster of optical microscopes. We have found that the robustness of IP networking systems allows a high level of fault tolerance in a wide range of settings important to the project, such as local use in isolated field research, or remote control over long distances. Furthermore, the rapid adoption of web APIs by developers all around the world means that an unprecedented range of resources are available for implementation in the microscope’s software, significantly lowering the barrier for community contributions.
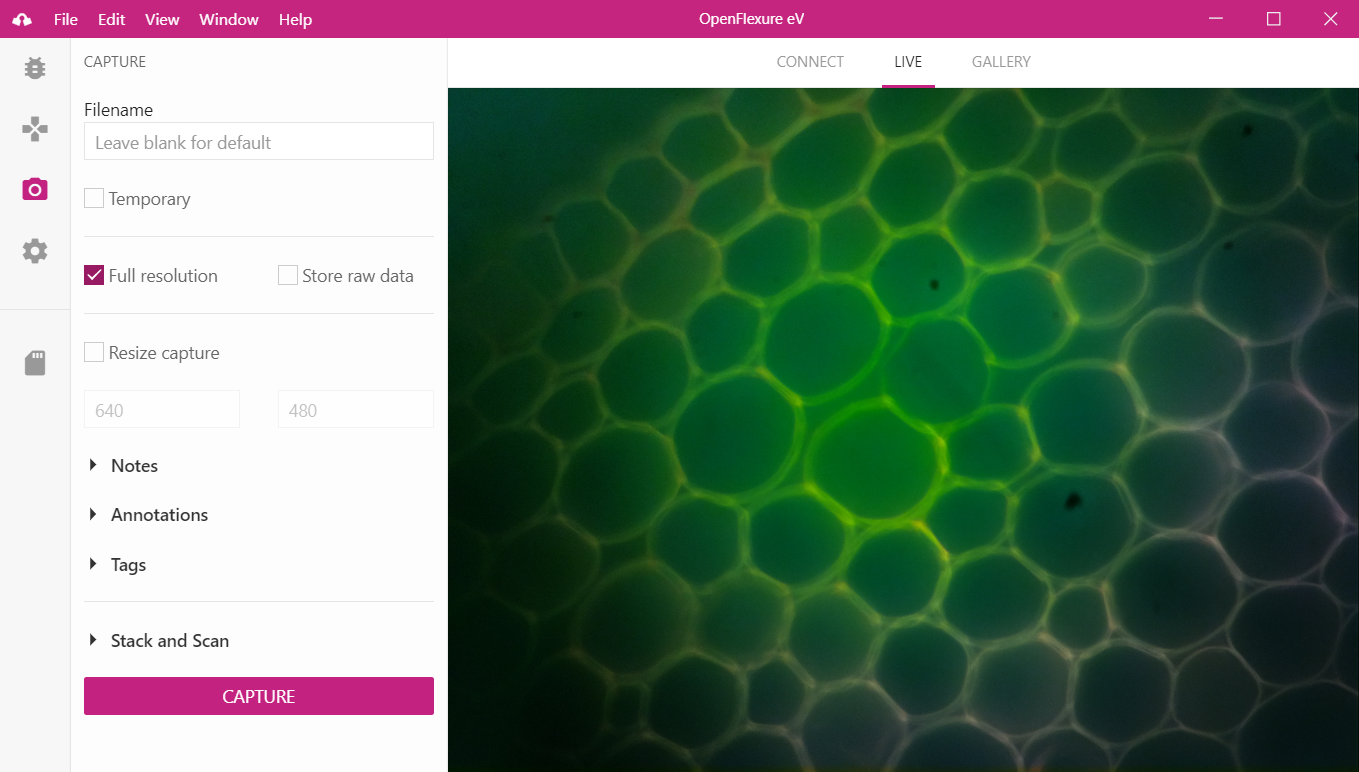
Figure 2 Graphical user interface for the OpenFlexure Microscope. The interface can be either installed as an application, or accessed from a web browser. The interface can be used to list and control a cluster of microscopes on the network, with interfaces components for all basic control (movement, capture, camera settings) as well as for microscope plugin functionality. A live camera stream (pictured) provides immediate feedback to users, while a gallery component enables per-microscope remote data management and transfer.
Our software stack handles automatic device discovery over any common network, using the Multicast DNS standard[2]. Microscope capabilities are presented using the W3C “Thing Description” standard[3], and interaction is handled by a web API conforming to the established “representational state transfer” (REST) architecture[4]. User extensions, adding functionality to the microscope, can be written in Python, and describe graphical user interfaces to allow simple use for end-users and non-programmers (Figure 2).
By developing a software stack based on these standards, and designed from the start to be extensible, we are building a strong global community across research, education, and healthcare, where every contribution can directly benefit all applications around the globe.
- References
[1] J T Collins et al, “Robotic microscopy for everyone: the OpenFlexure Microscope” bioRxiv (2019)
[2] S Cheshire and M Krochmal, “Multicast DNS” RFC Editor (2013)
[3] S. Kaebisch and T. Kamiya, “Web of Things (WoT) Thing Description” (2020)
[4] A. Keränen, M. Kovatsch, and K. Hartke, “RESTful Design for Internet of Things Systems” Internet Engineering Task Force (2019)
[5] The authors gratefully acknowledge funding from EPSRC (EP/P029426/1, EP/R013969/1, EP/R011443/1), the Royal Commission for the Exhibition of 1851 (Research Fellowship for RWB), Royal Society (URF\R1\180153, RGF\EA\181034)