Comparative transmission electron microscopic study of WT and VMP1-deficient Chlamydomonas
- Abstract number
- 1214
- Event
- European Microscopy Congress 2020
- DOI
- 10.22443/rms.emc2020.1214
- Corresponding Email
- [email protected]
- Session
- LSA.2 - Dynamic interactions in cells, organoids, tissue and entire organisms
- Authors
- Dr Eugenia Maximova (1), Dr Hezi Tenenboim (1)
- Affiliations
-
1. Max Planck Institute of Molecular Physiology
- Keywords
authophagy, Chlamydomonas, transmission electron microscopy, VMP1
- Abstract text
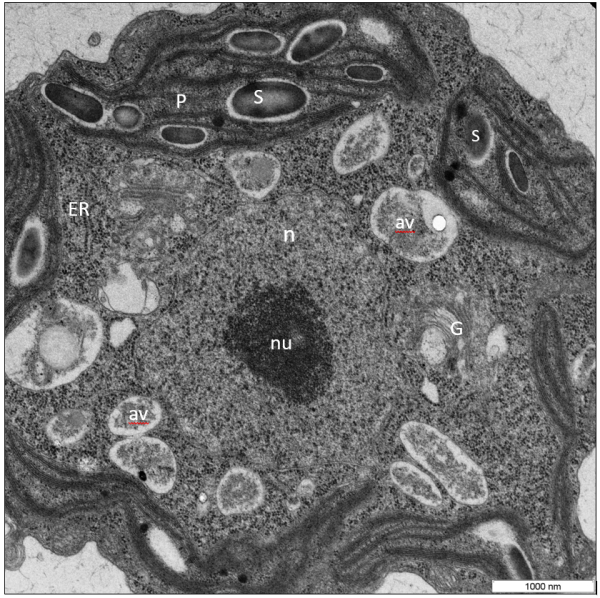
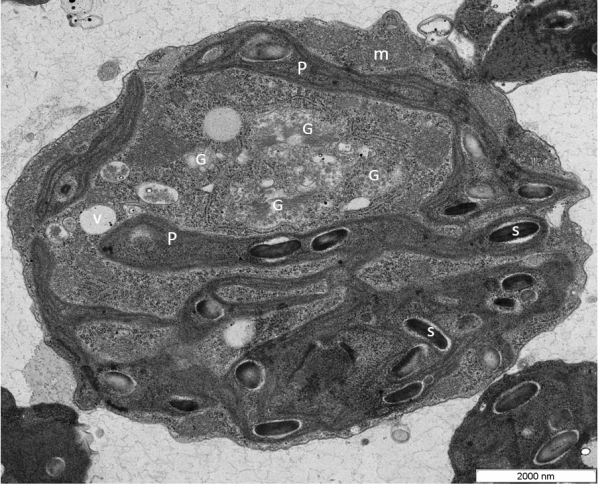
VMP1 (Vacuole Membrane Protein 1) is a transmembrane protein with homologues in all eukaryotic kingdoms, with fungi being a notable exception. It has been shown to be located in several cellular compartments: the Golgi apparatus [1], the endoplasmic reticulum, autophagosomes, and the plasma membrane. In this work we identified a VMP1 homologue (henceforth named CrVMP1) in the green alga Chlamydomonas reinhardtii. The cell-wall-deficient C. reinhardtii strains CC-4350 (cw15 302) and UVM11 were used in this study. Cells were grown in Tris-acetate-phosphate (TAP) medium and were cultured in 22°C, with light intensity of 85 μmol/m2/sec, while shaking at 125 rpm, and, unless stated otherwise, under continuous light. For transmission electron microscopy, Chlamydomonas cells were fixed for 3 h at 4°C with 2% (v/v) glutaraldehyde in 0.1 M sodium cacodilate buffer (pH 7.4), then post-fixed for 1 h in 1% (w/v) OsO4. Dehydrated progressively in series of ethanol and were embedded in Spurr's low-melting epoxy resin. Samples were degassed and cured at 60°C for 24 h. Sections (50 nm) were examined with an energy-filtering transmission electron microscope (EFTEM) at 120 kV (Zeiss, Germany). Electron microscopy revealed in the mutant (Fig.1) elongated mitochondria up to threefold greater than WT mitochondria, with markedly dilated cristae; disorganized and misshapen chloroplasts with numerous protrusions; autolysosomes in WT (Fig.2) often contained electron-dense matter but not in the mutant - these matter-containing vacuoles are most likely autolysosomes, and their absence in the mutant may indicate a defect in autophagy; abnormally numerous (3–4 per cell) Golgi apparatus compared to almost always one in WT (although many C. reinhardtii strains often exhibit two apparatuses, our UVM11 cells seldom did) and excessively large starch granules (Fig.1). The latter two phenomena are known to be linked: plants tend to accumulate starch when their Golgi system is inhibited or disrupted [2]. The resulting images revealed numerous irregularities in our mutants and concurred with the light microscopy observations: many cells displayed defective cytokinesis and grotesque cell morphologies (Fig.2). In this study we generated VMP1-deficient Chlamydomonas and subsequently identified both a novel role for VMP1 and a new gene involved in Chlamydomonas cell division and autophagy. CrVMP1-deficient cells exhibited disrupted cytokinesis and aberrant morphologies. Cell-biological and biochemical evidence suggests impaired autophagy: mutant cells accumulated triacylglycerides and formed enlarged starch granules; they were devoid of autolysosomes although WT cells showed many; and they showed enlarged mitochondria. In addition, mutants under expressed several key genes involved in cell-cycle regulation and autophagy.
References:
[1] Dusetti NJ et al., Biochem Bophys. Res Commun 2002, 290:641-649.
[2] Hummel E.,et al., J Exp Bot 2010, 61:2603-2614.
Figures.
Figure 2.The Ultrastructure of a Chlamydomonas VMP1-deficient cells. Cells are oriented with the anterior near the top as determined by location of the nucleus, starch granules. The chloroplast has an ameboid shape, making protrusions. Golgi stacks appear to have points in continuity with ER (arrows). Golgi apparatuses are very prominent and are 5-6 per section and vesicles are numerous and swollen.Figure 1. Transmission electron microscope image of Chlamydomonas WT (UVM11). Chloro-amyloplasts has typical structure: the starch grains are bigger size; lamellae are oriented along the axis of the plastid, stacked thylakoid membranes are prominent within the chloroplast. Golgi apparatus presented on section by 1-2 per section. Nucleus placed in the central position. In the cell are viewed autophagic vacuoles (av).
Abbr.: av, autophagous vacuole; e, eyespot; G, Golgi apparatus; m, mitochondrion; n, nucleus; ER, endoplasmic reticulum; P, plastid; S, starch granule; t, thylakoid membrane; py, pyrenoid; v, vacuole.
v
Abbr.: av, autophagous vacuole; e, eyespot; G, Golgi apparatus; m, mitochondrion; n, nucleus; ER, endoplasmic reticulum; P, plastid; S, starch granule; t, thylakoid membrane; py, pyrenoid; v, vacuole.