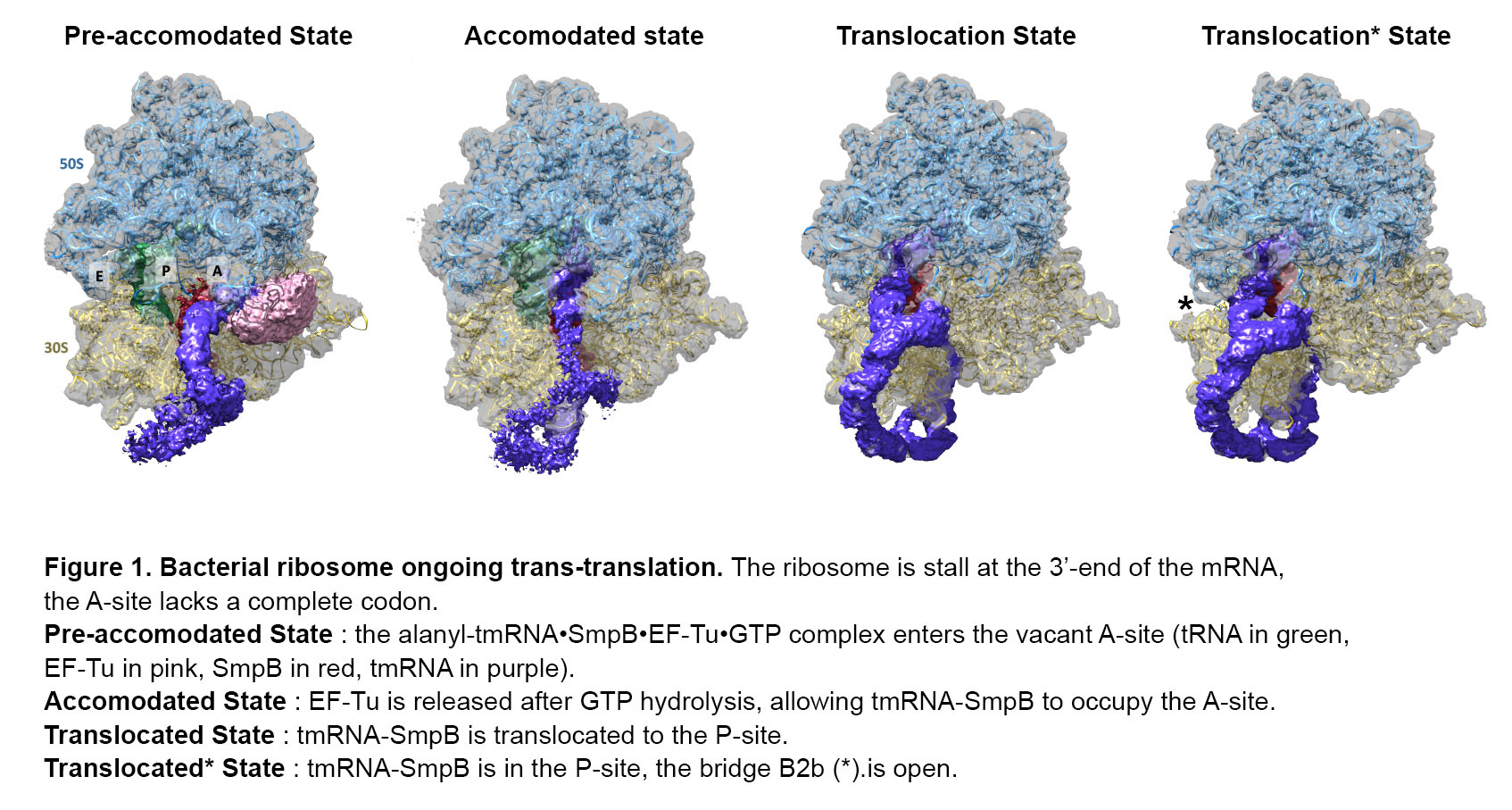
Deciphering trans-translation by cryo-electron microscopy: motions and interactions of tmRNA and SmpB inside the ribosome
- Abstract number
- 649
- Event
- European Microscopy Congress 2020
- DOI
- 10.22443/rms.emc2020.649
- Corresponding Email
- [email protected]
- Session
- LSA.3 - Applications for imaging sub-cellular events at high resolution
- Authors
- Charlotte Guyomar (1), Gaetano D'Urso (1), Emmanuel Giudice (1), Sophie Chat (1), Daniel Thomas (1), Reynald Gillet (1)
- Affiliations
-
1. IGDR UMR 6290 CNRS UR1
- Keywords
cryo-electron microscopy, ribosome, SmpB, tmRNA, trans-translation
- Abstract text
In bacteria, trans-translation is the primary quality control mechanism to rescue ribosomes stalled during translation. This key process is universally conserved and plays a crucial role in the viability and virulence of bacteria, thus making it a very attractive target for the development of new antibiotics. Two main actors drive the system: the unconventional transfer-messenger RNA (tmRNA) and its protein partner named small protein B (SmpB). tmRNA acts as a key molecule during trans-translation, it has the properties of both mRNA and tRNA. Its tRNA-like domain (TLD) resembles the upper part of a tRNA, and is associated with SmpB. The second important domain is the messenger-like domain, an internal open reading frame that encodes for a tag specifically recognized by proteases.
Various structural analyses, including cryo-electron microscopy, have widely contributed to understand the different trans-translation steps. They mainly described the TLD moving through the ribosome during pre-accommodation, accommodation and translocation [1,2,3]. However questions still remain unsolved: how SmpB interacts with the whole tmRNA? What are the motions of the different domains of tmRNA during the different stages of trans-translation?
Here we present a high-resolution study of trans-translation in E. coli, derived from our recent cryo-electron microscopy data. Thanks to the acquisition of data from the latest generation cryo-EM machines in Heidelberg, and Strasbourg (iNEXT, FRISBI), we have obtained several trans-translation complexes at resolutions close to 3.5 Å with an unprecedented signal level on the different domains of tmRNA. We describe precisely some new movements and interactions of tmRNA and SmpB with various ribosomal components during the steps of pre-accommodation (pre-ACC), accommodation (ACC) and translocation (TRANS) to the P-site (figure 1). We obtained the first high-resolution map in cryo-microscopy, describing the pre-ACC state, with a visualisation of the pseudo-node circle of the tmRNA. We are able to accurately describe the interactions of the H5 helix of tmRNA with the C-ter tail of SmpB, as well as the interaction between the body of SmpB and the TLD/H2 helix of tmRNA, for the pre-ACC and ACC states. As a novelty, we have captured a new intermediate state between the P-site TRANS and the E-site TRANS. We describe the opening of a bridge (B2b), that permits the TLD, the H2 and SmpB propeller to pass to the E-site.
Our results are in agreement with the latest published structural data [3]. However, they provide a deeper understanding of some key steps in trans-translation. Since trans-translation is specific to the bacterial world, each step should represent a very attractive target for the development of new broad-spectrum antibiotics. Thus, the ability to solve such related structures close to the atomic resolution is a prerequisite for their development.
- References
[1] Valle M, et al. Science, 300 (2003), p.127-130
[2] Weis F, et al. EMBO J, 29 (2010), p. 3810-3818
[3] Rae CD, Gordiyenko Y, Ramakrishnan V. Science, 363 (2019), p. 740-744