Multiplane Microscopy, a tool to study fast 3D dynamics at the microscale
- Abstract number
- 1400
- Event
- European Microscopy Congress 2020
- DOI
- 10.22443/rms.emc2020.1400
- Corresponding Email
- [email protected]
- Session
- LST.10 - Lightsheet illumination/detection strategies to yield higher speed, higher resolution and higher throughput in Bioimaging
- Authors
- Mr. Boris Louis (1, 2), Mr. Sergey Abakumov (1), Mr. Johannes Vandaele (1), Prof. Hiroshi Masuhara (3), Prof Ivan Scheblykin (2), Dr. Roger Bresoli-Obach (1), Dr. Rafael Camacho (1), Prof. Susana Rocha (1), Prof. Johan Hofkens (1)
- Affiliations
-
1. KU Leuven
2. Lunds Universitet
3. National chiao Tung University
- Keywords
3D microscopy, dynamics,microrheology, single-particle tracking, trapping
- Abstract text
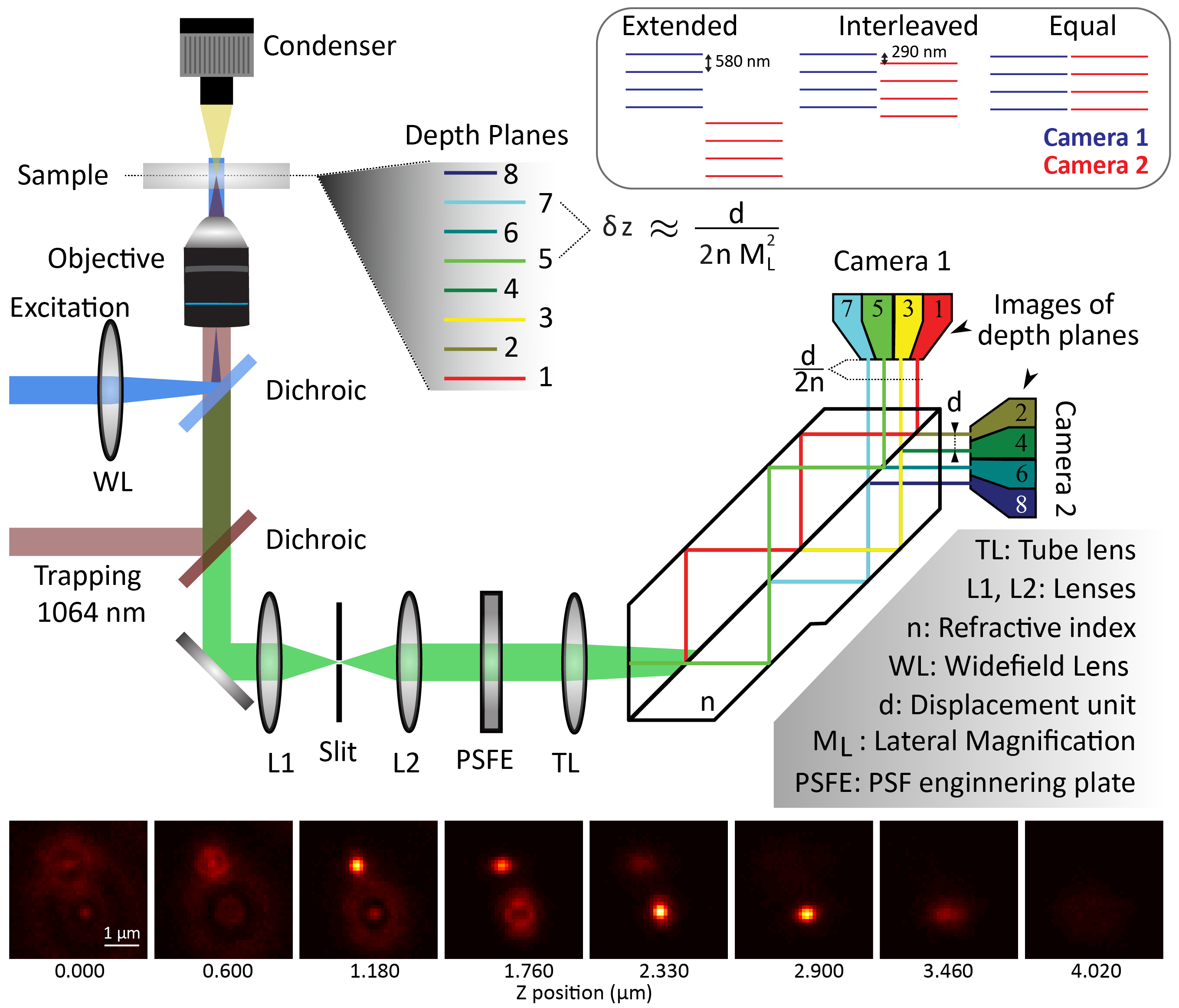
One of the advantages of fluorescence microscopy over high resolution method such as electron microscopy or atomic force microscopy is its ability to probe dynamics. Many fluorescence-based methods such as Fluorescence correlation spectroscopy, Image correlation spectroscopy or particle tracking allow to obtain dynamics information. However, most of these methods have only been developed for one or two dimensions except tracking that has seen some development in three-dimensions. This is due to the fact that acquiring three-dimensional data typically needs z-stacking which takes time and is therefore incompatible with fast dynamic measurements. To overcome this issue, we developed a multi-plane microscope. Our home-built system is largely based on the design described by Descloux et al. [1] and is essentially a wide-field setup to which we added a proprietary prism after the tube lens. The prism acts as a multiple beam-splitter which split the image in 8 while inducing a slightly different optical path to each of these 8 images leading them to be focused at different depth. The 8 images are collected in two groups of 4 by 2 cameras (see Figure 1).
By design, our multiplane microscope (see Figure 1) has a distance between imaging planes of 580 nm. Using hardware synchronization and two sCMOS cameras, we are able to record 50x50x5 um at a speed > 200 frames per second. This allows us to probe a wide range of dynamics, which are important in various fields ranging from molecular biology to optical trapping. We have developed two main approaches to extract dynamics from the 3D images acquired, the first one is 3D particle tracking while the second is 3D differential dynamic microscopy.
In particle tracking we have reduced the processing time significantly by obtaining the X,Y,Z coordinate in a single one-dimensional fit. This is thanks to the use of the phasor method by Martens et al [2] which obtain X,Y coordinate without the need for fitting. Then we used the phasor magnitude which is very sensitive to the signal-to-noise (and thus, the focus) and fit the z dependence with a 1D gaussian.
Differential dynamics microscopy (DDM) was developed by Cerbino and Trappe 2008.[3] and is based in the analysis of the differences between different temporal images in the Fourier space, which allows to identify the different frequencies of the involved dynamics. The main advantage of DDM over tracking experiments is that is not necessary to track any structure, so more complex shape and/or concentrated samples can be analysed. Additionally, it can easily be applied to any type of microscopy modality (Bright-field, dark-field, fluorescence). Finally, to our best knowledge DDM was never applied to 3D data.
Herein, we demonstrate the feasibility of the implementation and compare the DDM results and applicability with single-particle tracking. To test and compare the performances, we have tracked motor-induced motion on fluorescent beads embedded in a gel sample. Additionally, we have studied different sizes of beads (200, 500, 1000 nm diameter) in water and show that we could extract correct values of viscosity for the 3 sizes of 1cP (at 20C). We also show that DDM works better for high concentration as tracking will often loose the trace. Finally, we demonstrate our tracking method by following particle to an optical trap and the DDM by extracting dynamic of mitochondria inside cells.
Figure1: Multiplane Widefield Microscope: Schematic of the presented multiplane wide-field microscope. The trapping laser, the PSF engineering plates and the proprietary prism are the 3 main components that differ from common widefield microscope. Inset shows the different alignment of camera that are achievable with the system. Images of 2 beads in the 8 planes acquired simultaneously, the difference in focus can clearly be observed.
- References
[1] A. Descloux et al., ‘Combined multi-plane phase retrieval and super-resolution optical fluctuation imaging for 4D cell microscopy’, Nat. Photonics, vol. 12, no. 3, pp. 165–172, Mar. 2018, doi: 10.1038/s41566-018-0109-4.
[2] K. J. A. Martens, A. N. Bader, S. Baas, B. Rieger, and J. Hohlbein, ‘Phasor based single-molecule localization microscopy in 3D (pSMLM-3D): An algorithm for MHz localization rates using standard CPUs’, J. Chem. Phys., vol. 148, no. 12, p. 123311, Mar. 2018, doi: 10.1063/1.5005899.
[3] R. Cerbino and V. Trappe, ‘Differential Dynamic Microscopy: Probing Wave Vector Dependent Dynamics with a Microscope’, Phys. Rev. Lett., vol. 100, no. 18, p. 188102, May 2008, doi: 10.1103/PhysRevLett.100.188102.