Real-Time Data Acquisition and Analysis
- Abstract number
- 554
- Event
- European Microscopy Congress 2020
- DOI
- 10.22443/rms.emc2020.554
- Corresponding Email
- [email protected]
- Session
- DHA.3 - Machine assisted acquisition and analysis of microscopy data
- Authors
- Marco Oster (1), Andreas Wisnet (1), Dominic Tietz (1), Hans Tietz (1)
- Affiliations
-
1. TVIPS GmbH
- Keywords
Camera CMOS Data In-Situ Processing Real-Time
- Abstract text
With the availability of high-speed image acquisition systems, experiments in transmission electron microscopy get more and more data intensive. This becomes especially problematic if data analysis is not able to keep up with data acquisition, as the required high-speed disk space for short-term data storage is always limited. For example, our state-of-the-art fiber optically coupled CMOS camera system TemCam XF416 [1] is capable of recording 4kx4k frames digitized in 16 bits at a rate of 48 frames per second, producing 1.5GB of data each second.
While in principle such data rates can be recorded with readily available components, special imaging properties can be exploited to either transform recorded information into a more compact or more accessible representation. For example, in the so-called single-electron counting mode, the normalized pixel data are discriminated for single electron events and summed up in a drift-corrected fashion framewise into a single image. Typically, the data present in such a single 4kx4k frame may have originated from tens of gigabytes of raw data.
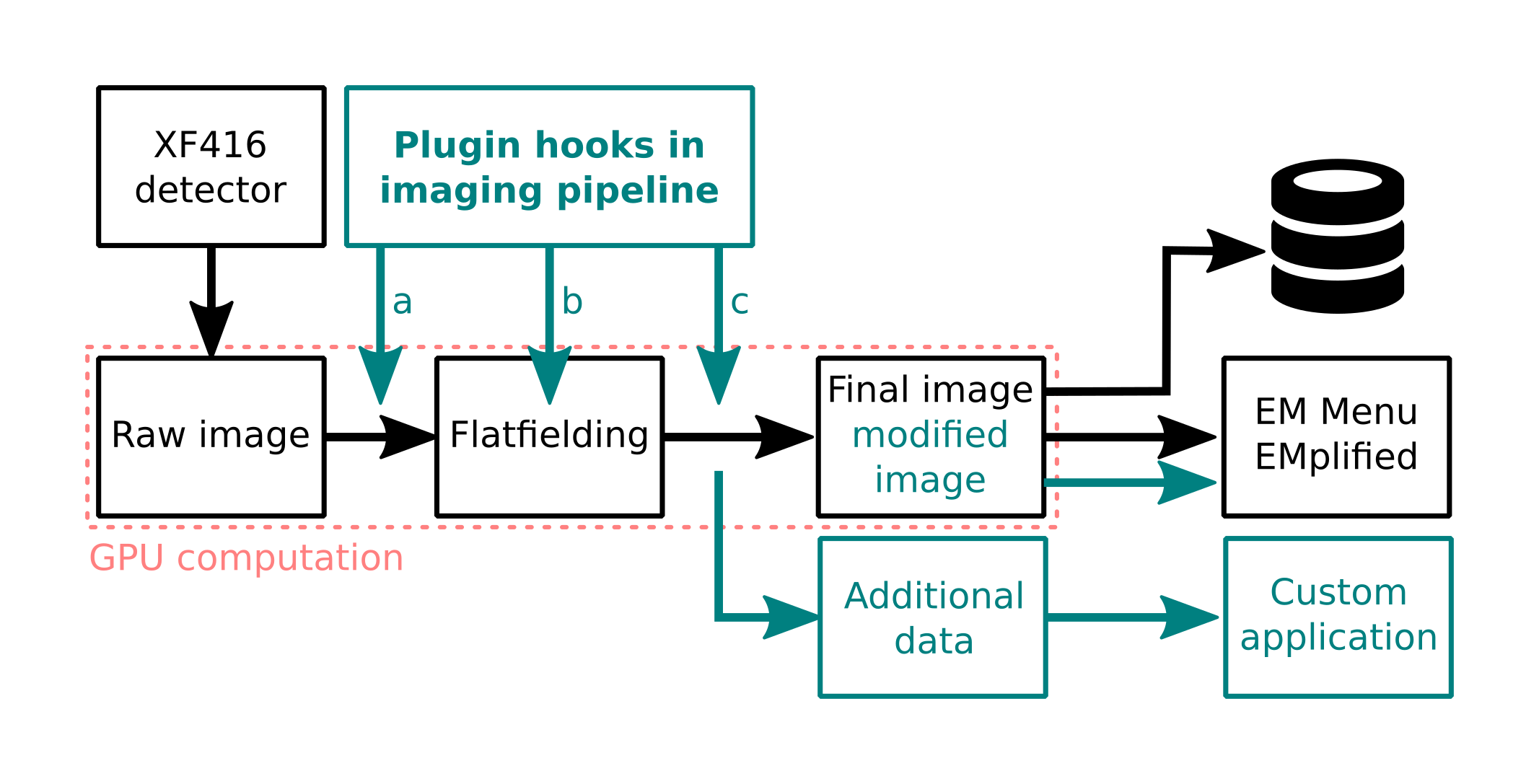
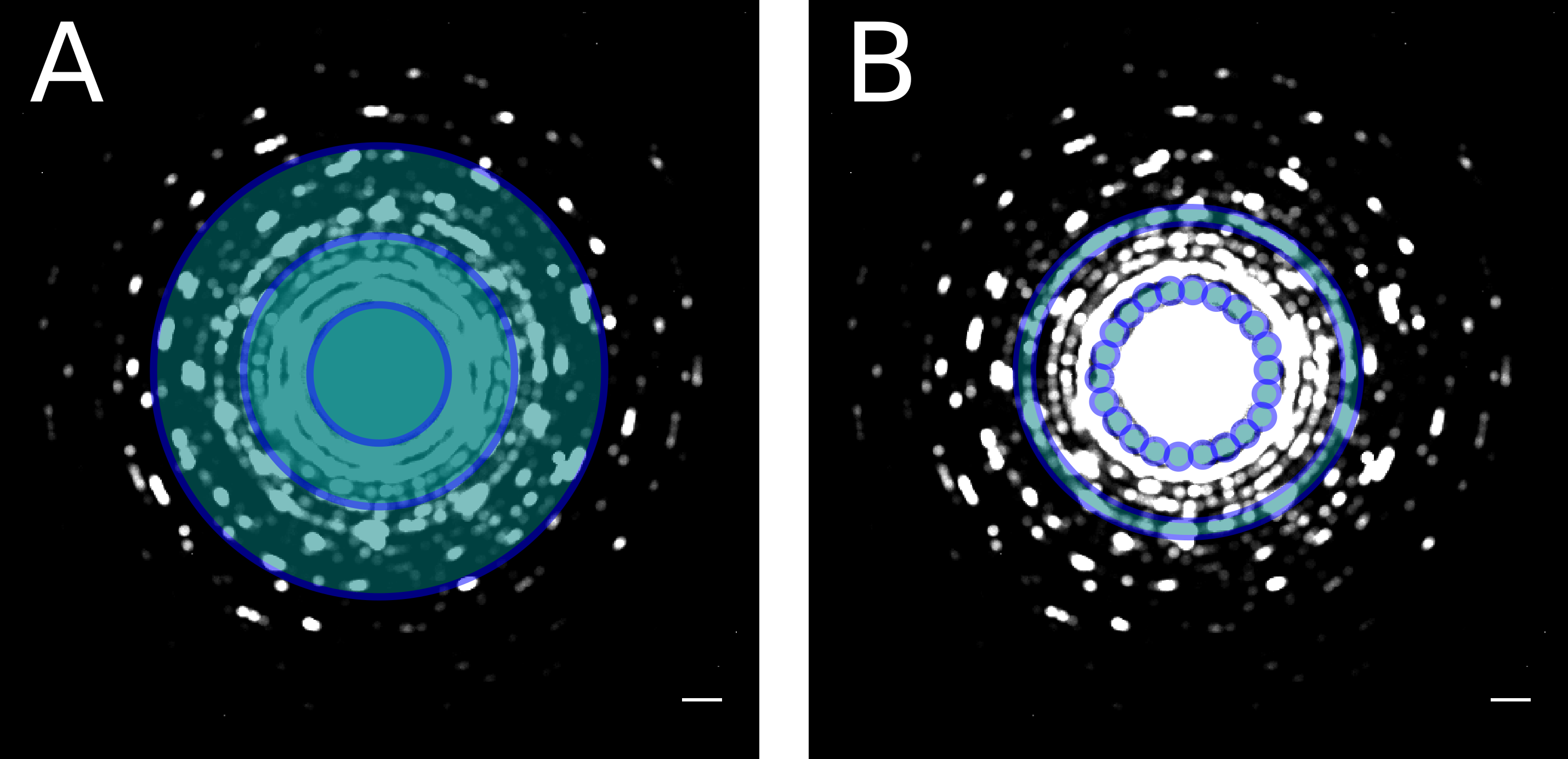
Generalizing from this special case, we present our GPU-based image processing pipeline, which processes all image data from the detector in real-time. Using a medium-sized GPU, dark and gain corrected 4kx4k images are available in GPU memory after a processing time of only 3ms, much faster than the system’s minimal frame time of 20ms. With the remaining processing time, further analysis can be carried out. This enables advanced capabilities, such as real-time drift-correction, single electron counting, high-speed extraction of spectra for STEM-EELS experiments [2], definition of virtual detectors for synchronized 4D-STEM experiments (Figure 2) and many more.
Our flexible image processing pipeline (Figure 1) can be tapped in several places, allowing to work on i.e. raw, normalized, or drift-corrected images. It is flexible enough to provide custom data channels, for example computing a signal corresponding to the apparent image shift and using that signal to excite the microscope’s deflectors or piezo stages to compensate for implementing a hardware drift correction scheme.
Some experimental setups also lend themselves to real-time data analysis: For instance, in 4D STEM experiments using a precessed nano beam electron probe, the measured data can be reduced to the center of mass positions of diffraction disks and their integrated intensity, which drastically reduces the required amount of data while still keeping all the relevant information for subsequent analysis.
On request, the API necessary for implementing such a real-time analysis scheme can be made available to customers, which allows users to integrate their custom image processing algorithms directly into our acquisition pipeline for best support of their experiments.
Figure 1: Image processing pipeline can be tapped in several places to accommodate any kind of real-time data analysis during the experiment.
Figure 2: Possible virtual detector configurations for 4D STEM experiments.
- References
[1] M Oster, R Ghadimi and H Tietz Microscopy and Microanalysis (2018), 24(S1), 896-897. doi:10.1017/S143192761800497X
[2] F Kahl, V Gerheim, M Linck, H Müller, R Schillinger and S Uhlemann. Advances in Imaging and Electron Physics Including Proceedings CPO-10 (2019), 212, 35 – 70 doi:10.1016/bs.aiep.2019.08.005