U-net segmentation of tubular structures from live imaging confocal microscopy: Successes and challenges
- Abstract number
- 418
- Event
- Virtual Early Career European Microscopy Congress 2020
- Presentation Form
- Submitted Oral
- DOI
- 10.22443/rms.emc2020.418
- Corresponding Email
- [email protected]
- Session
- DHA.3 - Machine assisted acquisition and analysis of microscopy data
- Authors
- Kasra Arnavaz (2, 1), Assistant Professor Pia Nyeng (4, 1), PhD Jelena Krivokapic (1), Assistant Professor Oswin Krause (2), Professor Aasa Feragen (3, 2)
- Affiliations
-
1. Danstem, University of Copenhagen
2. Department of Computer Science, University of Copenhagen
3. DTU Compute
4. Roskilde Universitet
- Keywords
U-net segmentation, tubular structure, pancreas, live-imaging, confocal microscopy
- Abstract text
Summary
We study the problem of segmenting tubular structures in the pancreas from live imaging via confocal microscopy. The segmentation is difficult due to noise, making it challenging even for a human expert to distinguish structure from background. We present the results of the state-of-the-art U-Net and inspect its successes and failures. We discuss challenges in the manual annotation of the structure, the impact of these challenges on our choice of experimental design, and future directions for improving automatic segmentation.
Introduction
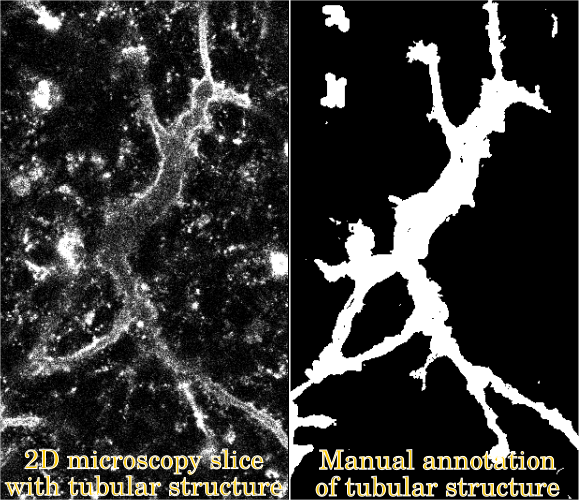
During embryonic development, the pancreatic tubes (Figure 1) grow from a web-like network with loops to a tree-like structure without loops. A thorough understanding of the mechanisms governing this morphology has yet defied developmental biologists [1], but observations indicate that the topological events happening during development of the tubular structure may be related to differentiation of beta cells nearby. However, confirming this quantitatively requires a binary segmentation of the tubes.
Figure 1: The segmentation problem.
While segmentation of other tubular structures such as airways or vessels is well studied, the pancreatic tubes are different in the sense that we have no prior anatomical knowledge about their structure. Moreover, the images are recorded with low signal to noise ratio, creating a challenging segmentation task. We present results of the state-of-the-art U-net [2] and discuss its successes and failures. In particular, we discuss challenges in the manual labeling of training data, as well as potential strategies for "optimally" labeling data.
Methods/Materials
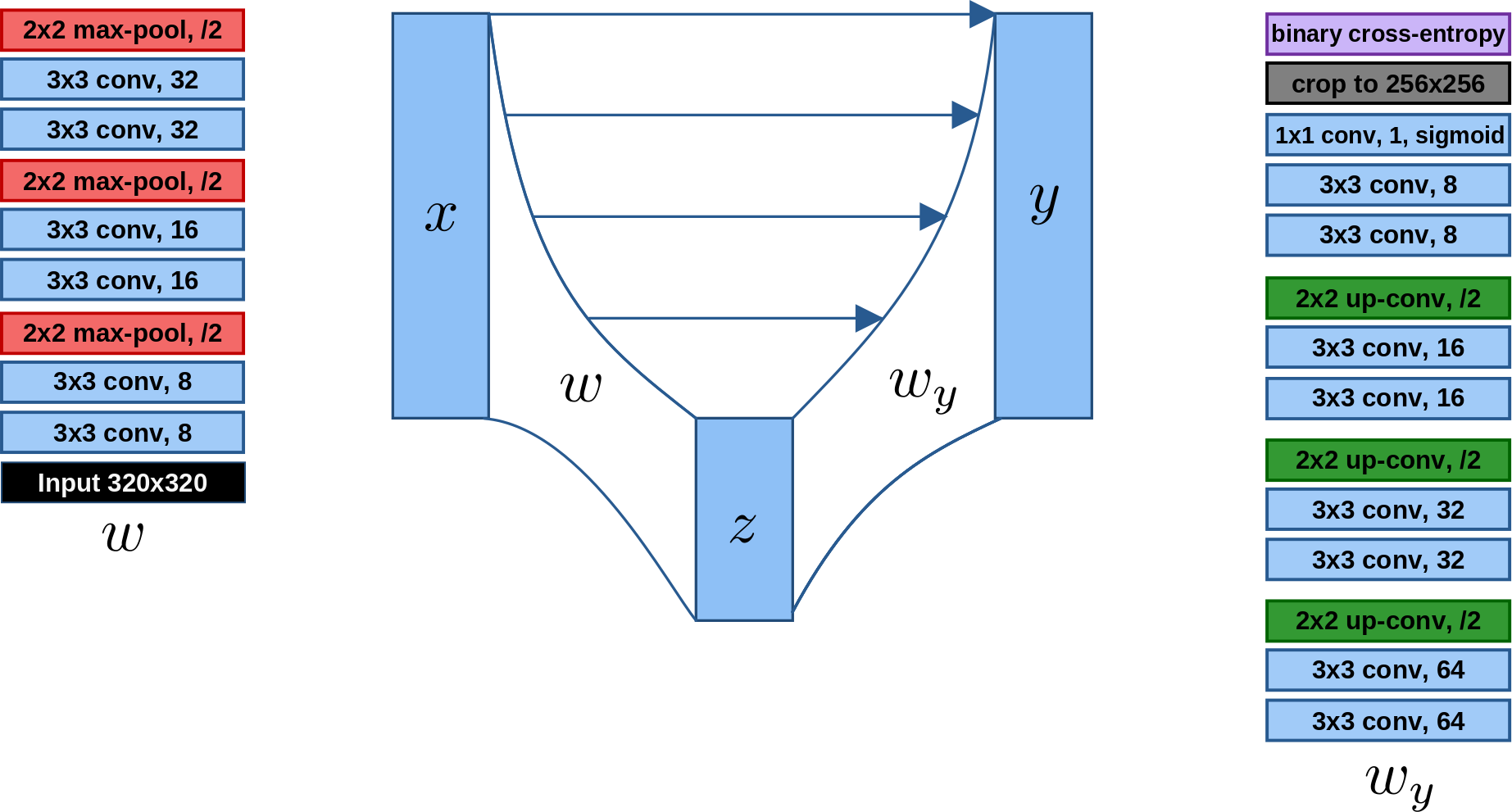
We implement the U-net (Figure 2) in Keras, using 3 sets of down- and up-sampling layers and binary cross entropy loss, and train it using the Adam Optimizer (beta_1 = 0.9, beta_2 = 0.999, lr = 10^{-5}) for 50 epochs.
Figure 2: The U-net.
Our dataset consists of 2D image slices extracted from 3D image stacks obtained from live imaging using confocal microscopy of the developing pancreas of mouse embryos imaged for 40-48 hours in vitro (see abstract submission from P Nyeng).
The 2D slices are aggregated as follows: A total of 6 3D movies with approximately 220 time-points each are split randomly into two sets with 3 movies each, to be used for training and testing, respectively. As parts of the movies have very high noise levels with no discernible structure, a smaller set of images is manually subselected by first randomly sampling, from each movie, two 3D images at random. These images have a resolution of 1024 x 1024 x D, where D varies between 27 and 40. Each of these images is divided into a 4 x 4 grid of patches in the x-y plane, each of size 256 x 256 x D. From this set of patches, a trained laboratory assistant manually removes all patches which are too dark and noisy to be reliably annotated. The remaining patches typically form connected parts of the x-y plane, leading to 1024 x 1024 x D images with ``forbidden regions" consisting of the removed patches.
Manual annotation: The training split consists of 6 3D images from 3 different movies with in total 174 2D image slices, where the tubular structure is fully annotated in a semi-manial fashion as follows: The 3D images were first processed in Imaris image processing software using Z-normalization, smoothing, volume rendering and filtering of the Muc1-mcherry signal. The resulting segmentations are manually edited by the laboratory assistant using the software Slicer. To validate on as diverse a dataset as possible, we select from the test split 10 3D patches at random, in which the tubular structure is annotated by the laboratory assistant, giving annotations for around 13% of the test split.
Image preprocessing: While the mean intensity per movie is close to zero, the variance differs noticeably. Thus, every time-point is stadardized by its own mean and standard deviation, and intensities are clipped to be at most 3 standard deviations away from the mean (which is zero).
Training on patches: The U-net is trained on image patches from the fully annotated slices in the training set, avoiding the forbidden regions.
Results and Discussion
On the test set, the U-net segmentation produces:
- a sensitivity/true positive rate, giving the proportion of true foreground segmented pixels out of ground truth foreground pixels, of 0.648
- a specificity/true negative rate giving the proportion of true background segmented pixels out of ground truth background pixels, of 0.987
- an accuracy, giving the proportion of correctly segmented pixels over all pixels, of 0.970
- a dice score, giving a similarity between the predicted segmentation and the ground truth segmentation, of 0.687 (optimal score is 1).
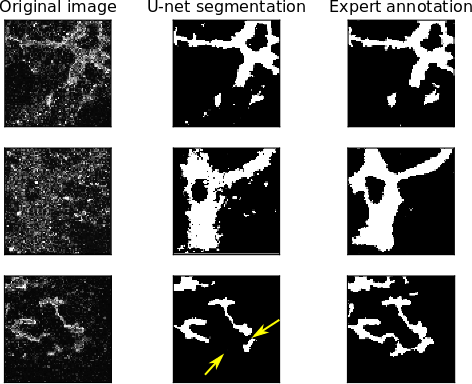
Example segmentations are shown in Figure 3. Note that the accuracy is high due to the class imbalance between foreground and easy-to-segment background. As shown by the sensitivity and specificity, however, the U-net is undersegmenting the foreground. This is problematic for two reasons. First, the expert annotation is already likely an undersegmentation, as the expert annotator is unlikely to annotate foreground unless she is fairly confident. Second, undersegmentation easily leads to disruptions in the topology of the segmented tubular network, as a few missing pixels can be enough to cut a connected tube into two or more pieces.
Figure 3: Examples of U-net performance compared to expert annotation. Note that undersegmenting the foreground can, in particular, greatly affect the topology of the segmented structure (bottom row).
Conclusion
We have demonstrated the performance of a U-net trained on patches from a subset of 2D slices from a small set of 4D microscopy live images. In light of the limited amount of training data, with limited amount of variation, the results are positive, although they also present challenges: The U-net is shown quantitatively to undersegment the tubular structure, which raises some concern regarding the correctness of segmented network topology.
In theory, this could be handled by increasing the hand-annotated data. In practice, however, manual labeling is expensive, difficult and time consuming. This leads to future research directions aiming to take advantage of our large amounts of unlabeled data.
- References
[1] HP Shih, A Wang, M Sander, Annu Rev Cell Dev Biol 29 (2013), p.81-105
[2] O Ronneberger, P Fischer, T Brox, MICCAI (2015), p.234-241
[3] The authors gratefully acknowledge funding from the Novo Nordisk Foundation under grant NNF170C0028360.