Uncovering cell dynamics in physiologically relevant 3D organoid and tumor models
- Abstract number
- 1065
- Event
- European Microscopy Congress 2020
- DOI
- 10.22443/rms.emc2020.1065
- Corresponding Email
- [email protected]
- Session
- LSA.2 - Dynamic interactions in cells, organoids, tissue and entire organisms
- Authors
- Gopi Shah (1), Jim Swoger (1)
- Affiliations
-
1. European Molecular Biology Laboratory
- Keywords
Cell dynamics, Embryoid, Light sheet microscopy, Live imaging, Organoids
- Abstract text
Recent trends in developmental biology and applied clinical research have introduced a variety of embryoid and organoid models that resemble the in vivo 3D environment. These physiologically relevant models such as mouse/human cell derived organoids provide a platform for visualising the dynamic behaviour of cells in various physiological and perturbed conditions, which is impossible to monitor in vivo. In recent years light sheet microscopy has proved to be an excellent technique for fast volume imaging of growing embryos of model organisms, owing to its ability to image large samples with minimal phototoxicity, and is becoming a tool of choice for imaging organoids as well. Organoids however are more sensitive to the imaging environment and susceptible to contamination, challenging long-term viability during the course of imaging. Moreover, they are quite variable in morphology as compared to embryos/tissues, requiring a larger number of samples to obtain a reliable understanding of the system. This poses challenges in terms of imaging throughput, data handling, visualisation of numerous big datasets and integration of information therein.
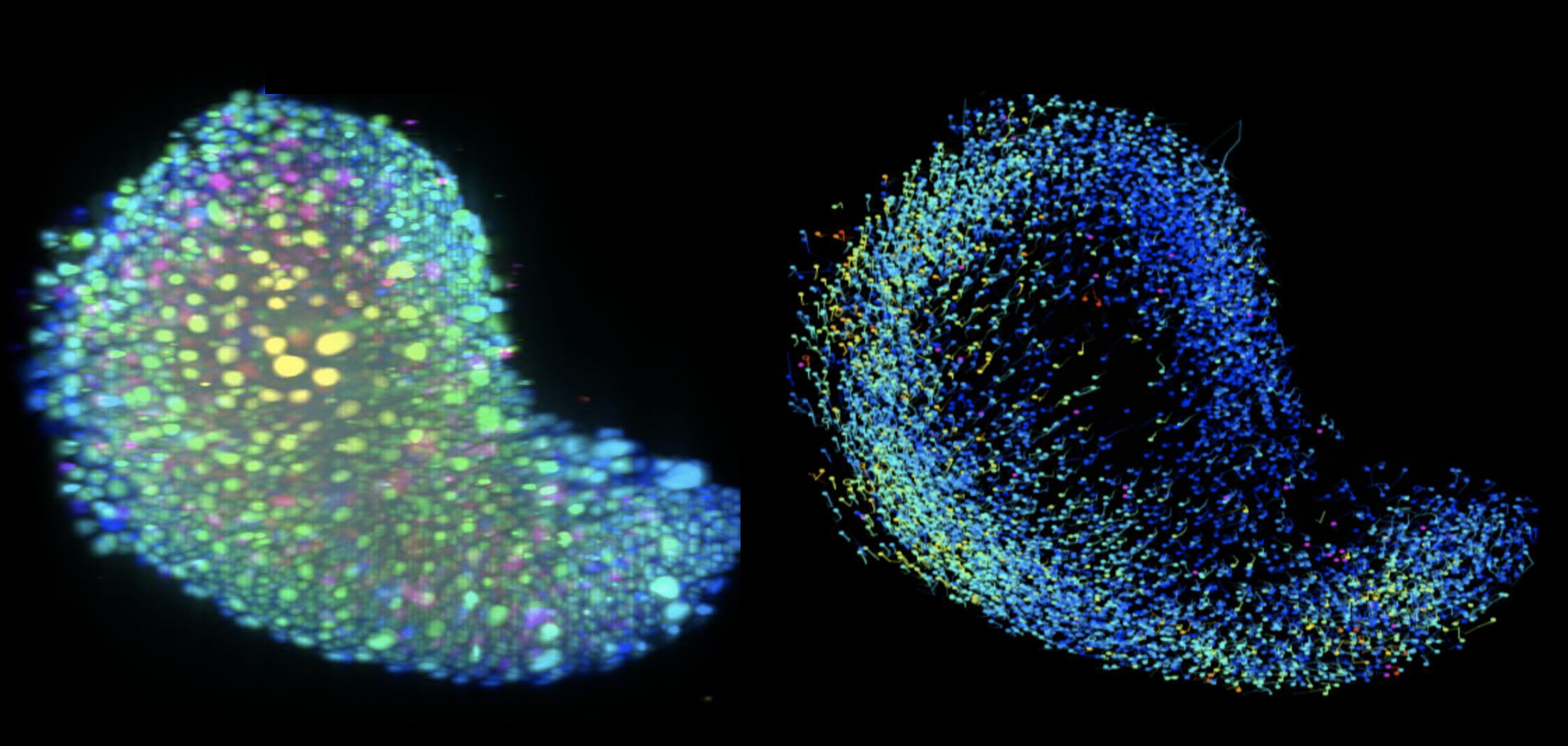
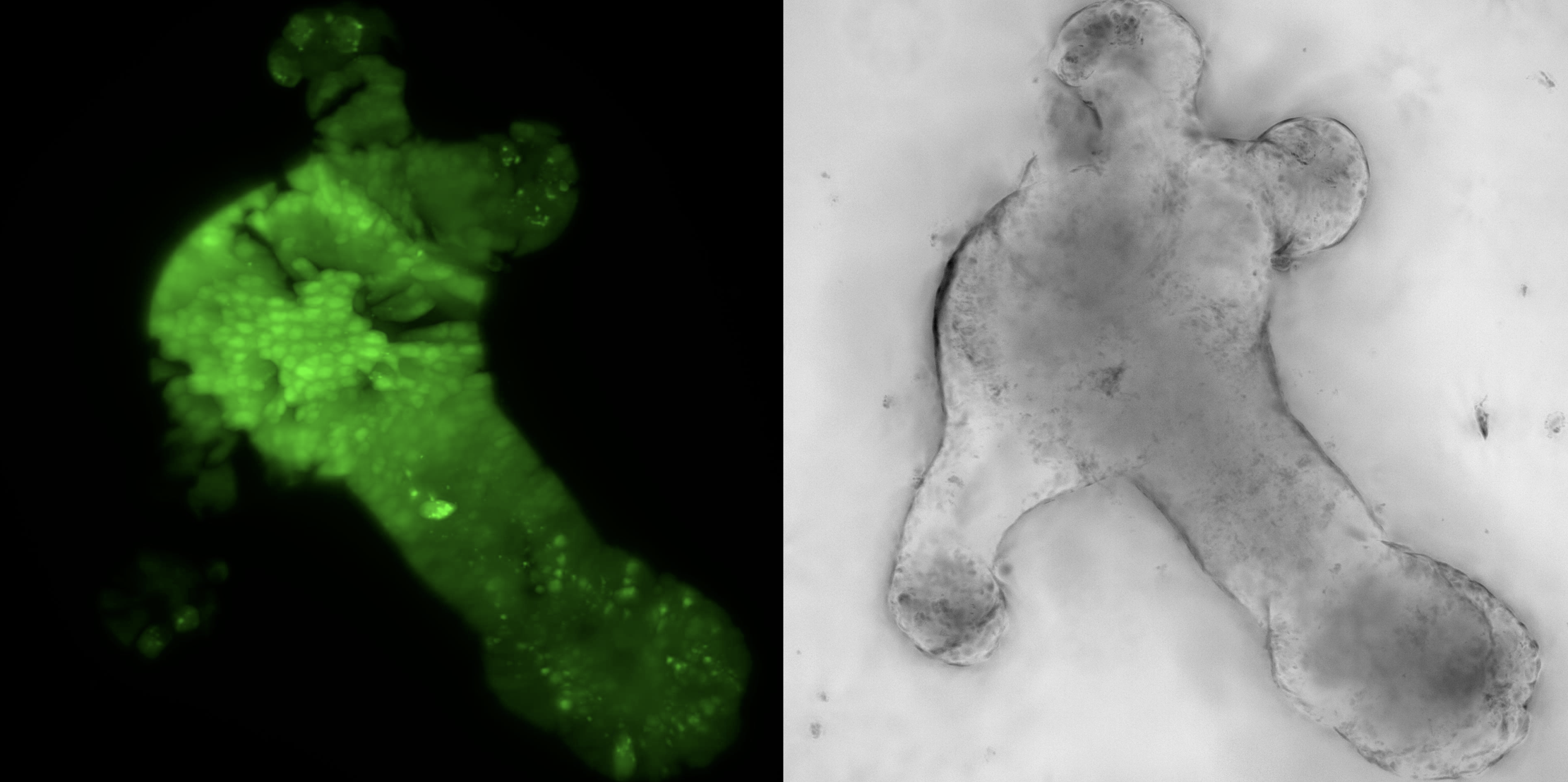
We are addressing these challenges by implementing robust incubation systems with regulated temperature, gas and humidity levels as well as sample carrier designs to support multi-sample imaging. I will discuss our workflows for time-lapse imaging of embryoid models (Figure 1) derived from zebrafish and mouse embryonic stem cells that begin as an aggregate of cells and develop in vitro to recapitulate events from embryonic development such as symmetry breaking and axis elongation. Further, I will discuss our experience with adult organoid systems derived from mouse intestine (Figure 2) and co-cultures of breast cancer cells to understand their interaction with the immune system. Our ability to visualise these biological processes in a dish enables us to identify, track and quantify the dynamics of individual cells and understand their interactions in a 3D environment. Through our on-going efforts for multi-sample data acquisition and visualisation, I would like to discuss the prospects that light sheet microscopy brings to the organoid community at large.
Figure 1: Light sheet microscopy images of an embryonic organoid undergoing elongation and its trajectories (Sample courtesy: Kerim Anlas, Trivedi lab EMBL Barcelona)
Figure 2: Light sheet fluorescence and bright field images of a mouse intestinal crypt organoid (Sample courtesy: Mariaestela Ortiz, Gilbertson lab, CRUK CI, Cambridge, UK)