Visualising the nanoscale dynamics and organisation of cellular membrane receptors
- Abstract number
- 1032
- Event
- European Microscopy Congress 2020
- DOI
- 10.22443/rms.emc2020.1032
- Corresponding Email
- [email protected]
- Session
- LSA.3 - Applications for imaging sub-cellular events at high resolution
- Authors
- Dr Aleks Ponjavic (2), Dr Alex Carr (1), Mr Edward Sanders (1), Mr Edward Jenkins (3), Dr Mafalda Santos (3), Dr James McColl (1), Professor Simon Davis (3), Professor David Klenerman (1), Dr Steven Lee (1)
- Affiliations
-
1. University of Cambridge
2. University of Leeds
3. University of Oxford
- Keywords
Light-sheet microscopy, Single-molecule imaging, Super-resolution microscopy
- Abstract text
Cells continuously interact with their environment to transmit signals and scan for infectious organisms at the cell membrane interface. Our understanding of these phenomena largely stems from single-molecule fluorescence imaging studies of membrane proteins in cells interacting with coated substrates. However, we have recently shown [1] that these substrates can artificially perturb the dynamics and organisation of membrane proteins in cells. I will introduce some novel fluorescence microscopy techniques that we have developed to enable the characterisation of proteins away from the perturbing glass substrate.
To study dynamics, we have developed a single-molecule light-sheet microscope that enables sensitive imaging of proteins in cells [2]. We have used this technology to observe the dynamics of proteins on the membrane of white blood cells, allowing us to study their diffusion in an environment without artificial perturbations.
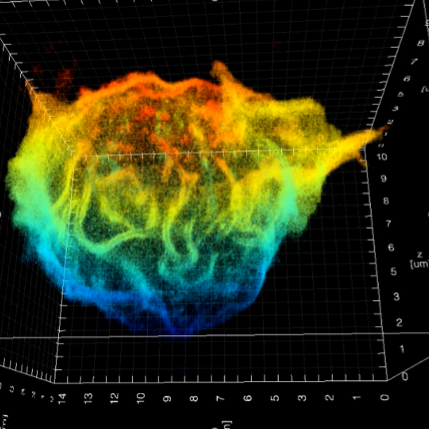
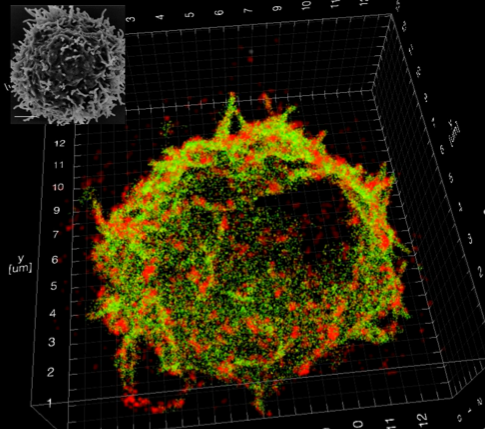
To study nanoscale organisation, we have applied double-helix point spread function (DHPSF) 3D super-resolution microscopy to image fixed cells. Most proteins visualised by super-resolution techniques organise into nanoclusters, which is believed to have an impact on signalling processes, but most work has been done in 2D and on artificial interfaces that we have shown are perturbative. Our new method allows us to map membrane protein distributions in 3D (Figure 1) as well as the cell membrane itself with a resolution of ~50 nm. By correlating membrane topography with 3D protein distributions, we can now explore whether receptors preferentially reside in the ~200 nm finger-like protrusions or if they are homogenously distributed, which will shed light on how organisation influences cellular function.
Figure 1: (Left): 3D super-resolution imaging of the entire membrane of a T cell @50 nm isotropic resolution. (Right): Quantitative super-resolution imaging of the T-cell receptor (red), with an overlay of the cell membrane (green). The inset shows an EM image of a T cell.
- References
[1] A M Santos*, A Ponjavic* et al, "Capturing resting T cells: the perils of PLL", Nature immunology 19(3) (2018), 203-205
[2] A Ponjavic et al, "Single-molecule light-sheet imaging of suspended T cells", Biophysical journal 114(9) (2018), 2200-2211